pyribs 0.4.0 introduces many new features, in particular in the archives. This post highlights some of the features and how to take advantage of them. For the full changelog, refer to the GitHub release.
Several changes are backwards-incompatible; these are listed at the beginning of their respective section.
Overview
Installation ¶
pyribs 0.4.0 supports Python 3.6-3.9. To install on pip, run:
pip install ribs[all]==0.4.0And on Conda:
conda install pyribs=0.4.0Instructions may differ depending on what distribution you installed previously. Refer to the Installation Selector for more details.
Archives ¶
Backwards-Incompatible: Rename index to index_0 in CVTArchive.as_pandas ¶
In previous versions, CVTArchive.as_pandas() returned a DataFrame with a column named index, since indices in the CVTArchive are only integers. For consistency with other archives, we have renamed this column to index_0.
Backwards-Incompatible: get_index() now public ¶
If you have not built your own archive, this change likely does not affect you.
To emphasize the meaning of the index in each archive, we have made get_index() public for all archives. Previously, this was a “protected” method known as _get_index().
Backwards-Incompatible: get_random_elite() and elite_with_behavior() return Elite ¶
Instead of a tuple of sol, obj, beh, get_random_elite() and elite_with_behavior() now return an Elite namedtuple. Elite contains sol, obj, beh, idx, meta, where idx is the index of the elite in the archive and meta is the metadata of the elite (more on that below). Furthermore, since Elite is a namedtuple, we recommend that new code use the following:
elite = archive.get_random_elite() # Or elite_with_behavior()
elite.sol
elite.obj
elite.beh
elite.idx
elite.metaNote that since Elite is a namedtuple, you can still use unpacking:
sol, obj, beh, idx, meta = archive.get_random_elite()However, the old unpacking will not work:
sol, obj, beh = archive.get_random_elite() # ERRORMetadata in archives ¶
In some cases, elites in an archive have arbitrary objects associated with them, such as generated images or dictionaries with extra statistics. With the introduction of metadata in pyribs, these objects can be easily stored with the elites. By default, elites store None as their metadata, but users may pass a list of metadata objects when calling tell() on emitters and optimizers. For instance, the following will associate the dict {"a": 1} with the first solution, None with the second solution, and the tuple (7, 8, 9) with the third solution:
optimizer.tell(
[1.0, 2.0, 3.0], # Objectives
[[1.0, 1.0], [2.0, 2.0], [3.0, 3.0]], # 2D BCs
[{"a": 1}, None, (7, 8, 9)], # Metadata - can be any object!
)Note that the old tell() interface still works, i.e. metadata is not required:
optimizer.tell(
[1.0, 2.0, 3.0], # Objectives
[[1.0, 1.0], [2.0, 2.0], [3.0, 3.0]], # 2D BCs
)As mentioned above, the Elite namedtuple contains meta for accessing metadata when retrieving an elite.
Furthermore, as_pandas() now has an include_metadata parameter, which adds a DataFrame column with the metadata of each elite. By default, this option is False so that the DataFrame can still be saved to a CSV.
Archive Statistics ¶
We have added the archive.stats attribute, which is an ArchiveStats namedtuple with useful statistics about the archive:
# Number of elites in the archive.
archive.stats.num_elites
# Proportion of bins in the archive that have an elite - always in the range
# [0,1].
archive.stats.coverage
# QD score, i.e. sum of objective values of all elites in the archive. This
# score only makes sense if objective values are non-negative.
archive.stats.qd_score
# Maximum objective value of the elites in the archive. None if there are no
# elites in the archive.
archive.stats.obj_max
# Mean objective value of the elites in the archive. None if there are no elites
# in the archive.
archive.stats.obj_meanArchiveDataFrame for as_pandas() ¶
Since as_pandas() splits arrays into individual components (e.g. solution_0, solution_1 and behavior_0, behavior_1), it was difficult to manipulate elites in a DataFrame. For instance, it was hard to access each solution vector when iterating over the rows of the DataFrame. It was also inconvenient to access the elite data in batch, e.g. “give me all the behavior values of the elites in this DataFrame.” To remedy this, as_pandas() now returns an ArchiveDataFrame.
ArchiveDataFrame adds several features to DataFrame’s which make it easier to manipulate elites. First, calling iterelites() on an ArchiveDataFrame returns an iterator which converts every row to an Elite namedtuple.
df = archive.as_pandas()
for elite in df.iterelites():
elite.sol
elite.obj
...Second, batch_X methods return the X of all elites in the ArchiveDataFrame. The methods currently available are:
batch_behaviors() # Array with behavior values of all elites.
batch_indices() # List of archive indices of all elites (each idx is a tuple).
batch_metadata() # Array with metadata of all elites.
batch_objectives() # Array with objective values of all elites.
batch_solutions() # Array with solutions of all elites.Note: After saving an ArchiveDataFrame to a CSV, loading it with pd.read_csv will return a DataFrame. To remedy this, pass the DataFrame from pd.read_csv to ArchiveDataFrame, as shown below.
# (Earlier)
archive.as_pandas().to_csv("file.csv")
# Loading from CSV.
from ribs.archives import ArchiveDataFrame
df = ArchiveDataFrame(pd.read_csv("file.csv"))Above all, since ArchiveDataFrame is still a DataFrame, all other Pandas features still work.
clear() method ¶
archive.clear() removes all elites from an archive.
__len__ ¶
len(archive) now returns the number of elites in the archive.
# (Initialize archive...)
archive.add(...)
print("Number of filled bins:", len(archive)) # Prints 1 in this case.__iter__ ¶
Iterating over an archive now yields every Elite contained in it. Importantly, for loops now work:
for elite in archive:
elite.sol
elite.obj
...Visualization ¶
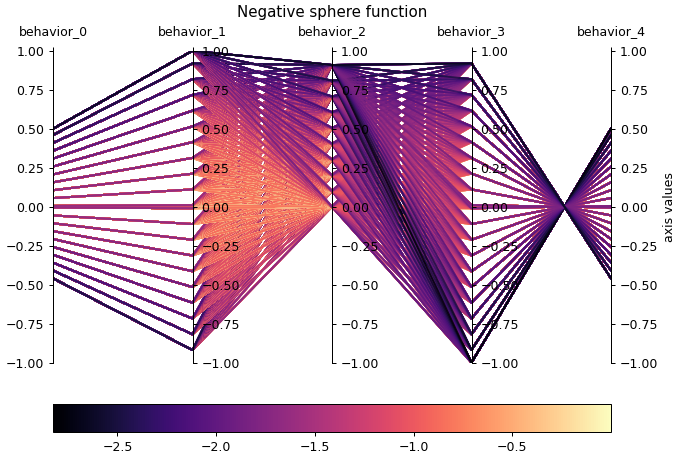
parallel_axes_plot for archives ¶
Thanks Nathan Dennler for contributing this feature!
We have added parallel_axes_plot for visualizing the elites of any archive in behavior space. In the parallel axes plot, each elite’s behavior values are displayed as a series of line segments connecting each value to the next. For instance, in the plot below, an elite with behavior values [0,5, 0.0, -0.5, 1.0, 0.0] would be shown with a line segment from behavior_0 = 0.5 to behavior_1 = 0.0, a line segment from behavior_1 = 0.0 to behavior_2 = -0.5, and so on. This plot can help with understanding the behavior space coverage of an archive, as well as the correlations between BCs (e.g. “is behavior_0 correlated to behavior_1?”).
Optimizers ¶
Support for emitters which return any number of solutions in ask() ¶
Previously, all emitters for an optimizer had to return the same number of solutions in their ask() method. Now, they may return any number of solutions – the number may even change across iterations.
Emitters ¶
Backwards-Incompatible: EmitterBase simplifications ¶
If you have not built your own emitter, this change likely does not affect you.
As the _rng and _batch_size / batch_size attributes are not required on all emitters, we have moved them out of EmitterBase and into children emitters. Since _rng is no longer in EmitterBase, we have also removed the seed parameter. Finally, we added an archive attribute (replacing the older _archive), which allows accessing the emitter’s archive.